Abstract
Background: Gene fusions function as therapeutic targets, impressively shown for e.g. BCR - ABL1 and PML - RARA . Hence, the need for a comprehensive genetic analysis as a basis for personalized medicine is increasing allowing to select treatment based on genotype and to provide markers for disease monitoring. At ASH 2016 we presented targeted RNA sequencing data from 38 patients with hematological malignancies harboring rearrangements one partner gene of which had been identified by standard diagnostic work up. Using targeted RNA sequencing the second partner gene could be identified in 22/38 cases. All rearrangements involving KMT2A (n=8), JAK2 (n=1), and RARA (n=1) were resolved. However, fusion transcripts were identified in only 4/9 ETV6 rearrangements and 8/19 RUNX1 rearrangements. In the present study a subset of these cases was further evaluated.
Aim: To evaluate the potential of whole genome sequencing (WGS) in combination with transcriptome (TS) sequencing to unravel the significance of rare translocations.
Patient cohort and methods: Five cases with unresolved ETV6 rearrangement and 4 cases with unresolved RUNX1 rearrangement were further analyzed by WGS (~90x coverage) and TS (~61m reads). WGS was performed using the Truseq DNA PCR-Free HT sample preparation kit and RNA was prepared using TruSeq Stranded Total RNA HT (with Ribo-Zero HMR) kit for TS. Library was prepared according to manufacture's protocol using fresh/frozen samples and sequencing performed on NovaSeq instruments in multiplexed runs (Illumina, San Diego, CA). Tumor-only sample analysis was conducted with BaseSpace (BS) WGS App with default parameters for alignment and subsequent (structural) variant calling. For further downstream analysis we loaded the data into BS Variant Interpreter software to filter down and prioritize variants of interest. TS analysis was performed using BS RNAseq App with default parameters to assess gene fusions and expression (Illumina, San Diego, CA).
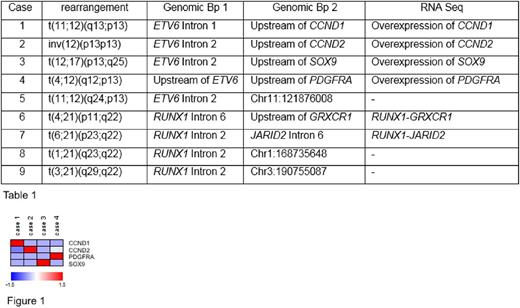
Results: In all 9 cases WGS detected the translocation predicted by chromosome banding analysis and the genomic breakpoints (Bp) were determined. Details are depicted in the table 1. In cases 1 to 4 the genomic breakpoints were located in introns 1 or 2 of ETV6 or upstream of ETV6 . The corresponding breakpoints were upstream of CCND1, CCND2, SOX9 and PDGFRA . By TS no fusion transcripts involving ETV6 were identified. However, in line with the genomic data an overexpression of CCND1, CCND2, SOX9 and PDGFRA was observed in the respective cases by TS (figure 1). Data suggest that an enhancer in the region of ETV6 is driving the overexpression of the respective genes. In case 5 the genomic breakpoint was located in intron 2 and no ETV6 fusion transcript identified by TS. Most likely the translocation leads to a stop codon and thus to haploinsufficiency of ETV6 .
In cases 6 and 7 a RUNX1 - GRXCR1 (RUNX1 exon 6 fused to GRXCR1 exon 2) and a RUNX1 - JARID2 (RUNX1 exon 2 fused to JARID2 exon 7) were detected by TS, respectively. The respective genomic breakpoints are depicted in the table 1. In cases 8 and 9 the breakpoints in RUNX1 were both located in intron 2. No RUNX1 fusion transcripts were detected by TS. Based on WGS and TS data the results of the RUNX1 translocations are most likely out of frame fusions and thus lead to loss of function of RUNX1 with an effect comparable to RUNX1 frameshift mutations.
In summary the combination of TS and WGS revealed 3 different mechanisms of ETV6 - and RUNX1 -rearrangements: 1. Overexpression of the partner gene, 2. Novel fusion transcripts, and 3. Loss of function of ETV6 or RUNX1 .
From the clinical point of view in case of PDGFRA overexpression therapy with a tyrosine kinase can be considered. The overexpression of CCND1 and CCND2 suggests that the mTOR inhibitor rapamycin might be a therapeutic option in these cases.
Conclusions and Perspectives: Whole genome sequencing in combination with transcriptome sequencing is a promising approach to unravel the pathogenetic role of rare rearrangements. This provides new options for therapeutic interventions and monitoring of minimal residual disease on a personalized level. Studies to evaluate the potential role of WGS and TS in a future diagnostic setting have been started to improve patients outcome based on a comprehensive genetic work-up allowing optimal treatment choice and monitoring of minimal residual disease.
Haferlach: MLL Munich Leukemia Laboratory: Employment, Equity Ownership. Nadarajah: MLL Munich Leukemia Laboratory: Employment. Walter: MLL Munich Leukemia Laboratory: Employment. Meggendorfer: MLL Munich Leukemia Laboratory: Employment. Kern: MLL Munich Leukemia Laboratory: Employment, Equity Ownership. Haferlach: MLL Munich Leukemia Laboratory: Employment, Equity Ownership.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal